SYLARAS (SYstemic Lymphoid Architecture Response ASsessment) is a preclinical research platform for the interrogation of systemic immune response to disease and therapy. The approach combines multiplex immunophenotyping with biological computation to transform complex single-cell datasets into a visual compendium of the time and tissue-dependent changes occurring in immune cell frequency and/or function in response to an arbitrary immune stimulus (e.g. tumor model, infectious or autoimmune disease, vaccine, immunotherapy, etc.).
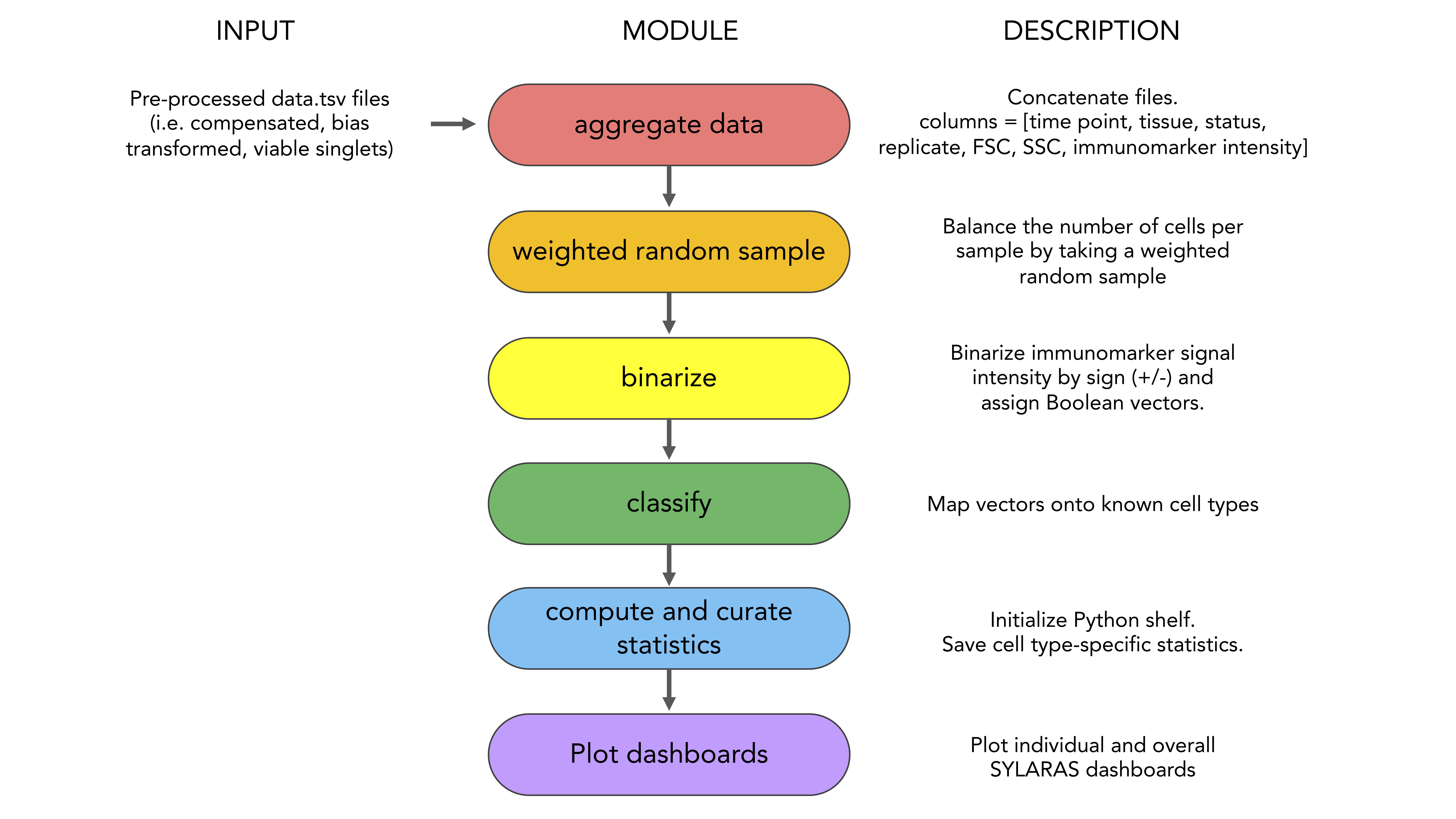
SYLARAS is deployed in three stages. In the first stage, longitudinal immunophenotyping data are collected from mouse lymphoid organs of test and control subjects in a high-throughput manner by multiplex flow cytometry. In the second stage, raw FCS files are spectrally compensated and filtered for viable cells before undergoing a systematic immune cell subset identification procedure. In the final stage, the pre-processed data are computationally analyzed using an open-source data analysis tool scripted in the Python programming language and run at the command-line of a personal computer. This leads to the generation of a set of data-rich graphical dashboards (1 per immune cell type) that together portray systemic immune response to a given experimental perturbation.
The ability to devise safe and effective medicines that elicit a targeted immune response to cancer or infectious disease hinges on a more integrated understanding of the cell and molecular mechanisms governing systemic immunology. A more holistic perspective can be achieved through detailed studies of immune structure and function using preclinical model systems that permit examination of immune response in multiple immune organs.
Although state of the art technologies for single-cell data acquisition allow for the rapid collection of expansive datasets pertinent to the study of systemic immune response, a companion data analysis tool capable of fully automating statistical analysis and data visualization is lacking. By leveraging a set of powerful Python-based computational libraries against bulky single-cell datasets spanning time, tissue, experimental perturbation, and biological replicate, SYLARAS overcomes the labor-intensity of manual cell subset identification, statistical analysis among subgroups, and visual display of single-cell data at scale.
Immune response programs can induce proliferation, migration, and differentiation of the biologically-specialized immune cell subsets comprising lymphoid tissue architecture. Although the molecular mechanisms underlying these changes may not always be clear, the resultant redistribution in immune cell frequency and inter-cellular correlation structure can be readily quantified by immunophenotyping: a technique for delineating the proportions of cell types based on differential antigen expression. SYLARAS uses this concept to infer changes in specific immune cell lineages and their network-level architectures secondary to an immune stimulus. The ability of SYLARAS to generate and display interpretable systemic immune response data makes it a broadly useful preclinical research tool.
When immunoreactivity is reported in binary terms (e.g. CDa+, CDb-, CDc+), the number of possible immunophenotypes specified by a panel of M antibodies is 2M. This exponential relationship makes comprehensive cell subset identification through serial gating cumbersome, time consuming, and, in many cases, non-commutative. SYLARAS addresses these drawbacks of manual gating through computation by programmatically assigning an immunophenotype "bar code" to each cell in the analysis.
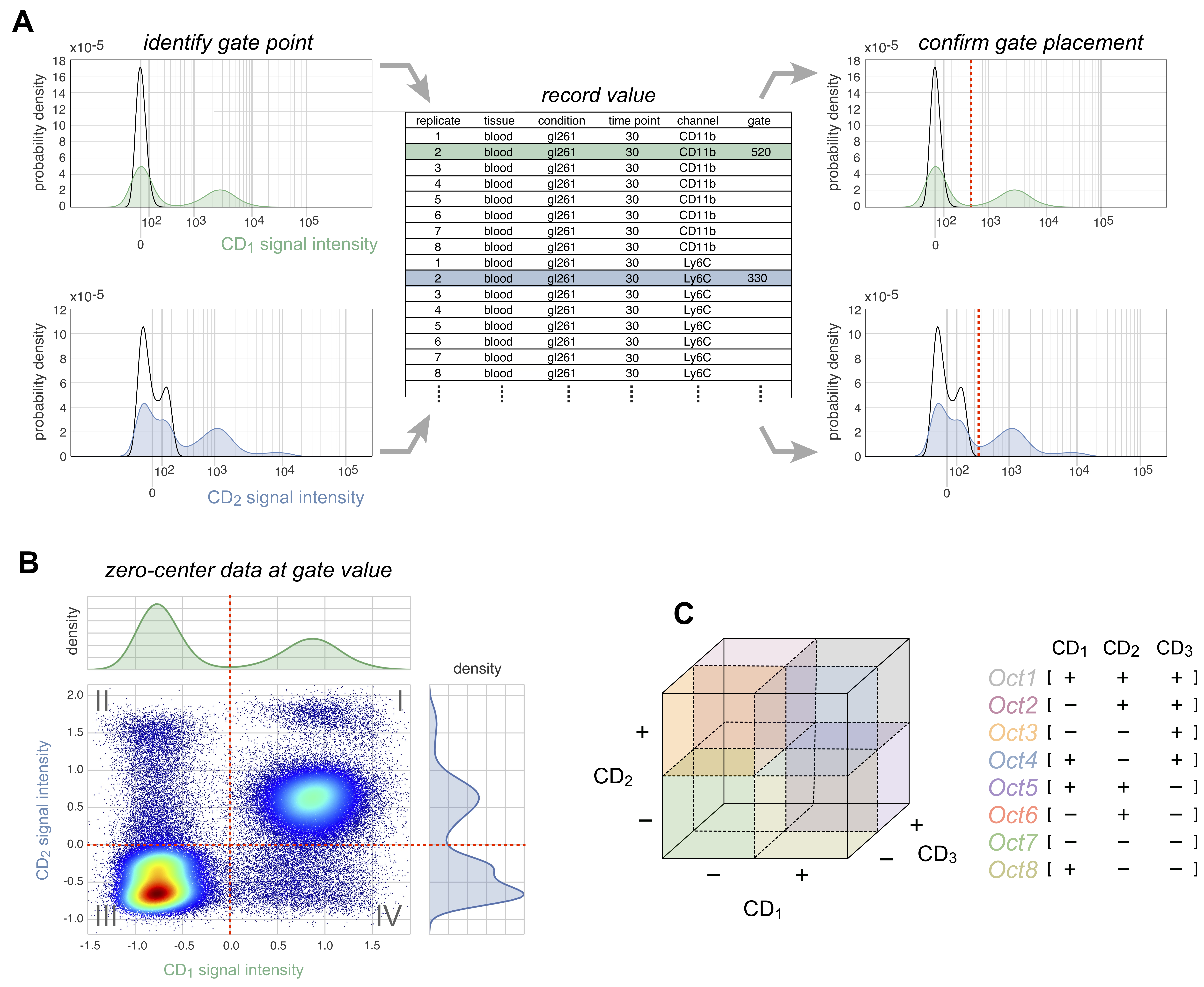
In the SYLARAS procedure, the interface between background and foreground signal is defined for each immunomarker by curating a set of one dimensional gates (histogram gates). The signal intensity distribution of unlabeled/isotype labeled cells is super-imposed on the immunolabeled distributions to aid in objective gate placement by revealing how autofluorescence and/or isotype control antibody binding compares to observed data ( panel a ).
The data then undergo a linear data transformation to center the predetermined gate values at zero. This causes background signal intensities to become negative valued and an M-dimensional Boolean immunophenotype to be programmatically assigned to each cell in the dataset ( panel b ). For example, a cell assigned the Boolean vector [1, 0, 1, 1, 0, 0, 0, 1, 1 ,0, 0] might correspond to the immunophenotype: CDa+, CDb-+, CDc+, CDd+, CDe-, CDf-, CDg-, CDh+, CDi+, CDj-, CDk-
In the case of a three immunomarker panel, the vectorization procedure can be conceptualized geometrically as the binning of cells across the 23 (= 8) octants of a 3-dimensional cube ( panel c ). Extrapolating the principle into higher dimensions (i.e. more immunomarkers), the vectorization procedure generalizes as the binning of cells across the 2M orthants of an M-dimensional hypercube.
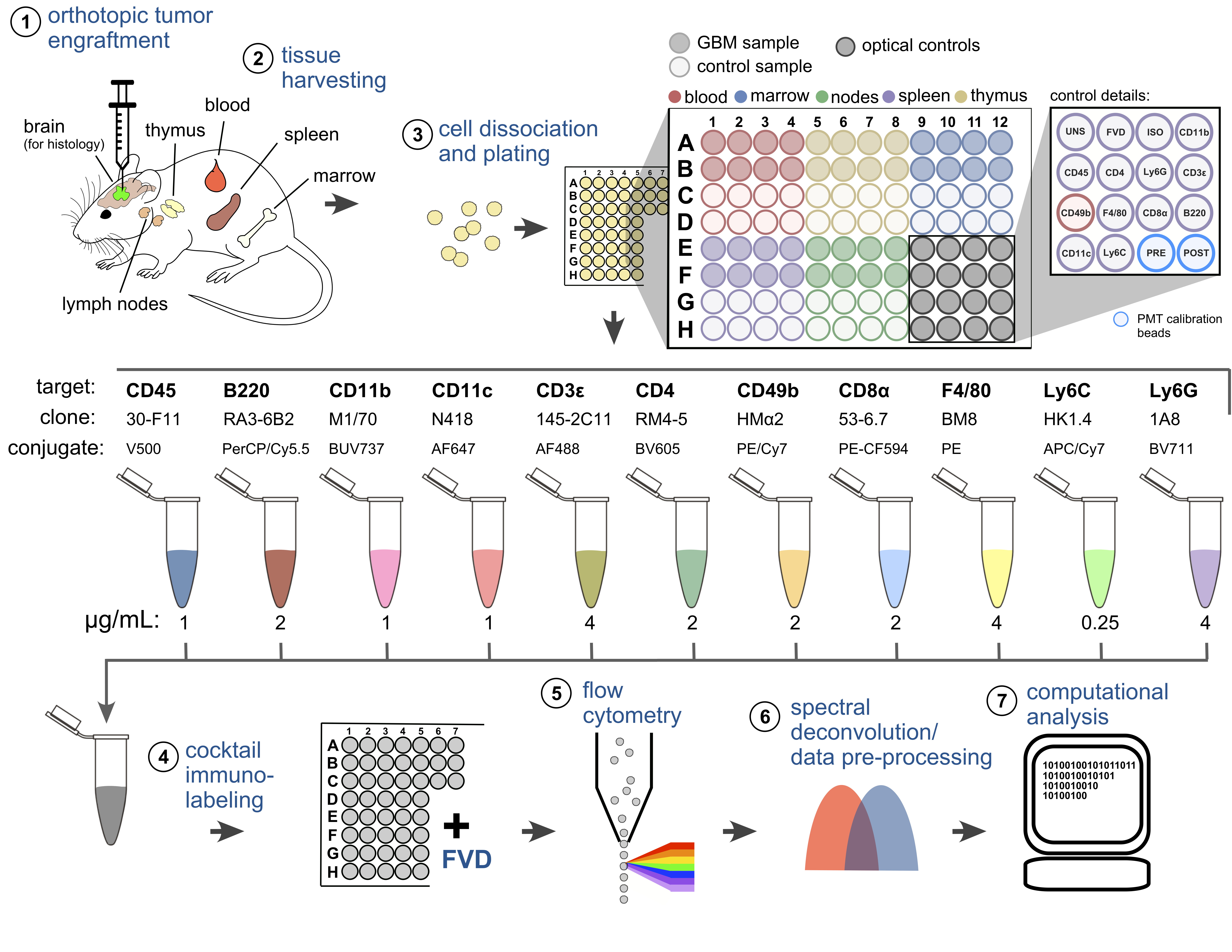
(1) An immune stimulus (glioblastoma brain cancer shown here) or control agent is administered to cohorts of age-matched mice. (2) Lymphoid tissues are harvested from mice of each treatment group at various time points after the onset of immune stimulation. (3) Tissues are processed into single-cell suspensions and plated in a 96 well V-bottom plate. (4) Cells are immunolabeled with a cocktail of fluorophore-conjugated antibodies then stained with a fixable viability dye (FVD). (5) Data are acquired by 12-color high-throughput flow cytometry. (6) Raw data are spectrally deconvolved and selected for viable singlets. (7) Pre-processed data undergo a computer-assisted gating procedure (see the "Cell Subset Identification" tab above for details) prior to computational analysis with SYLARAS software. Click here to download our experimental protocol.
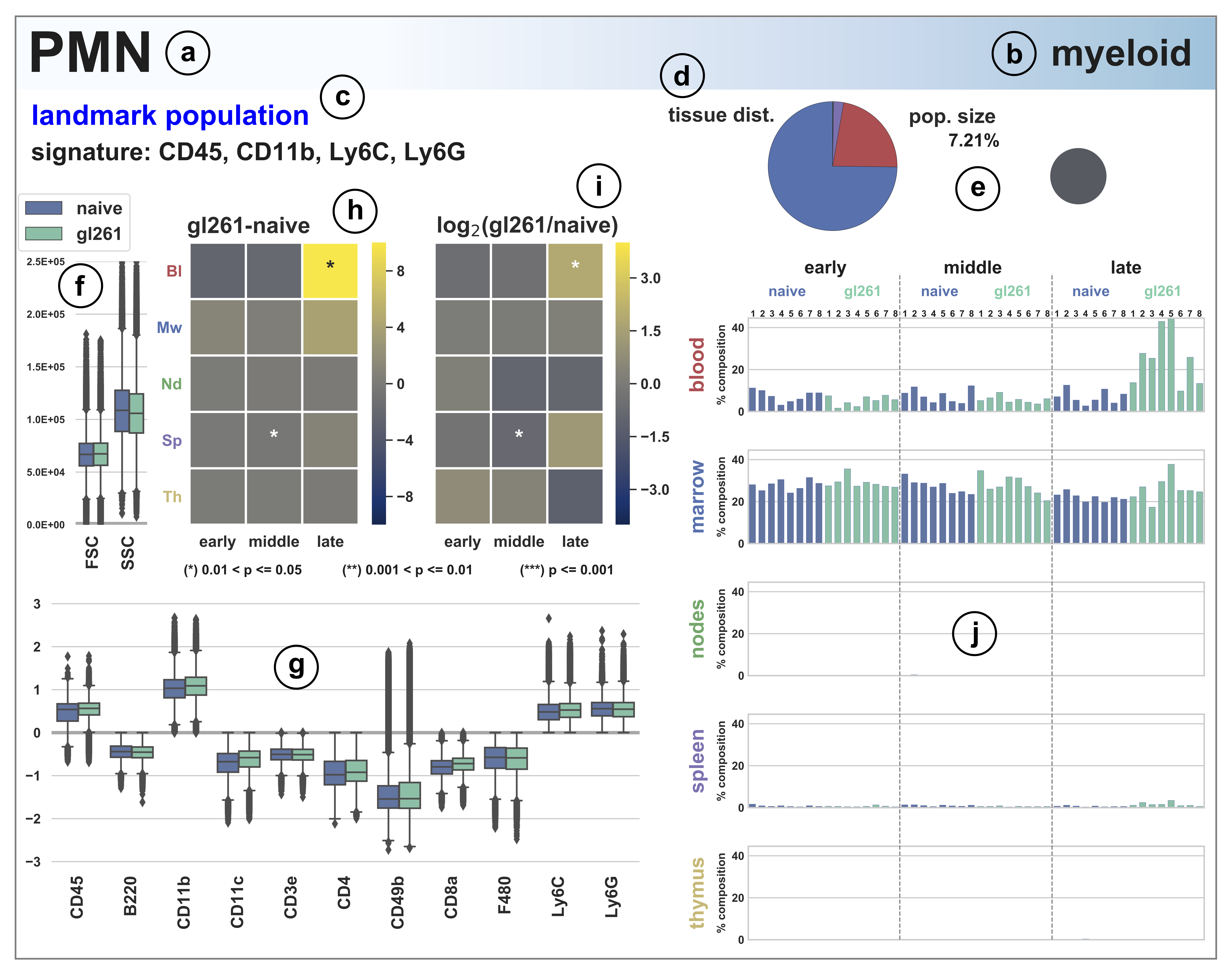
SYLARAS portrays longitudinal, multi-organ immunophenotyping data on a per cell type basis in a readily-interpretable dashboard layout. The example of polymorphonuclear (PMN) immune cells shown here was programmatically generated during a SYLARAS screen of the mouse immune response to syngneic, orthotopic glioblastoma(GBM) brain cancer. (a) brief alias; (b) lineage; (c) immunomarker signature indicating whether the immunophenotype corresponds to 1 of 14 major “landmark population”; (d) distribution of cells across 5 lymphoid tissues color-coded as in (h, i and j); (e) percentage of this cell type relative to all immune cells; (f) forward and side scatter (FSC/SSC); (g) Logicle-transformed, background-subtracted immunomarker signal intensity; (h and i) time and tissue-specific difference in mean percentage and log2 fold-change between GBM-burdened and mock-engrafted animals (n=8 mice/group), asterisks denote one of three levels of statistical significance; (j) contribution of this cell type (in percent) in each tissue across the study’s 48 mice.
Publications
(1) Baker et al. Cell Syst. 2020 Sep 23;11(3):272-285.e9. doi: 10.1016/j.cels.2020.08.001. Epub 2020 Sep 7, SYLARAS: A Platform for the Statistical Analysis and Visual Display of Systemic Immunoprofiling Data and Its Application to Glioblastoma. Click here to read the SYLARAS article.
(2) Baker et al. BioRxiv (2019) doi: https://doi.org/10.1101/555854, Systemic immune response profiling with SYLARAS implicates a role for CD45R/B220+ CD8+ T cells in glioblastoma immunology. Click here to read the SYLARAS preprint.
(3) Gregory J. Baker, Sucheendra K. Palaniappan, Stephanie H. Davis, Jodene K. Moore and Peter K. Sorger. Systemic lymphoid architecture response assessment (SYLARAS): Application to system-wide immunophenotyping of glioblastoma; Cancer Res July 2018 (78) (13 Supplement) 5670; DOI: 10.1158/1538-7445.AM2018-5670
(4) Gregory J. Baker, P.S. Thiagarajan, Sucheendra K. Palaniappan, Stephanie H. Davis, Jodene K. Moore and Peter K. Sorger. A flow-based immunoprofiling strategy for interrogating system-wide leukocyte composition in response to glioblastoma; Cancer Res July 1 2017 (77) (13 Supplement) 1678; DOI: 10.1158/1538-7445.AM2017-1678
Terms of Use
The SYLARAS algorithm is a copyrighted, open-source software intended for non-profit academic use.
Copyright (c) 2019 - President and Fellows of Harvard College. All rights reserved.
Developed by: Gregory J. Baker
Harvard Program in Therapeutic Science (HiTS), Harvard Medical School (http://hits.harvard.edu/)
Harvard University case number HU 7716 - A computational tool for systems immunophenotyping
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal with the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimers.
Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimers in the documentation and/or other materials provided with the distribution.
Neither the names of the Harvard Program in Therapeutic Science, HiTS, Harvard Medical School, Harvard University, the Harvard shield or logo, nor the names of its contributors may be used to endorse or promote products derived from this Software without specific prior written permission.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE CONTRIBUTORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS WITH THE SOFTWARE.
Funding
The SYLARAS project was supported by American Cancer Society Postdoctoral Fellowship PF-16-197-01-LIB to G.J.B, and by NIH/NCI grants P50-GM107618 and U54-CA225088 to P.K.S. and by the Harvard Ludwig Center.
Citation
Cite the SYLARAS approach to systems immunophenotyping and the use of its resources as: Baker et al. Cell Syst. 2020 Sep 23;11(3):272-285.e9. doi: 10.1016/j.cels.2020.08.001. Epub 2020 Sep 7. SYLARAS: A Platform for the Statistical Analysis and Visual Display of Systemic Immunoprofiling Data and Its Application to Glioblastoma.
Contact Us
Copyright © All rights reserved | This template is made with by Colorlib